Strand bias in STR motifs
CommentsSummary: Certain STR motifs are much more likely to occur on the coding strand in transcribed regions, suggesting STRs may play an important role in properties of transcription.
Short tandem repeats (STRs) are ubiquitous across the genome, with an average of several STRs per kilobase. Many of these STRs occur in transcribed regions (UTRs, exons, and introns). While most of these repeats are thought to have no functional consequences, some of them have been implicated in regulating transcription, suggesting that not all STRs are “neutral”.
If STRs indeed did not play any importance in transcription, one would expect that the strand on which a certain STR motif occurs shouldn’t matter. Therefore, if we look at for instance all repeats with an “AC” motif (this includes e.g. ACACACAC but also TGTGTGTG, since these are just the same motif on opposite strands), then we should be equally likely to see the “AC” repeat on the coding strand as we would to see the “TG” repeat on the coding strand. The same applies for other motif species: e.g. for “AAC” vs. “GTT” repeats, if STRs did nothing we’d expect to see “AAC” just as often as “GTT on the coding strand.
In their paper in PLOS One last year, Sawaya et al. indeed found strand biases for STR motifs around promoters. For example, they found for A/T and AC/TG motifs, there is a phase shift that occurs around transcription start sites: “A” and “AC” are more likely to be on the coding strand upstream of the TSS, whereas “T” and “GT” are more likely to be on the coding strand downstream of the TSS (see Figure 4 of their paper).
I decided to examine whether strand biases for specific motifs exist across transcribed regions compared to other parts of the genome. For each of commonly occurring STR motifs, I asked what percent of the time the “A” rich motif was on the coding strand for STRs occurring in:
- introns
- UTRs
- 0-2kb upstream of the TSS (to compare to Sawaya et al)
- 50-100kb upstream of the TSS (as a non-transcribed control region)
The results showed some pretty clear biases:
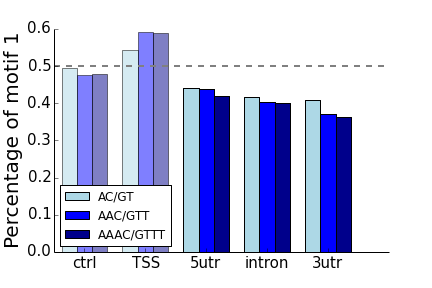
- AC/GT, AAC/GTT, AAAC/GTTT
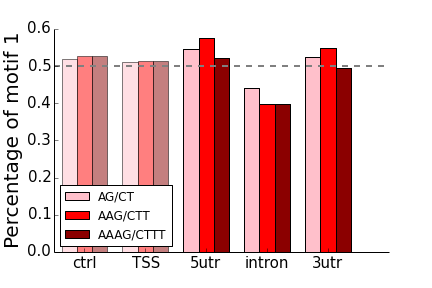
- AG/CT, AAG/CTT, AAAG/CTTT
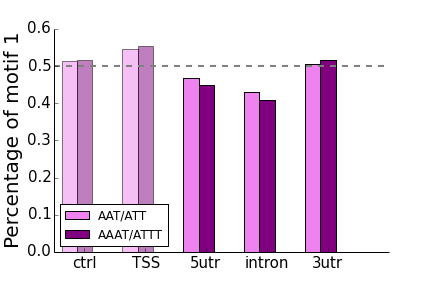
- AAT/ATT, AAAT/ATTT
Overall:
- All of the “control” non-transcribed regions of 50-100kb from the TSS have frequencies around 50%, as expected.
- The region around the TSS (0-2kb upstream) matches the bias seen by Sawaya (bias for AC vs. GT, no bias for AG vs. CT)
- In all cases for intronic repeats, the “T” rich motif of each pair is enriched on the coding strand compared to the “A” rich motif. For “AC”-type motifs, this enrichment is also present in the UTRs.
So the question: what drives the enrichment of “T” rich motifs on the coding strand in transcribed regions?. Several possible explanations:
- “GT”-type motifs can form hairpins using G-U base pairing in RNA, whereas AC-type motifs cannot. Therefore one hypothesis is that based on which strand the GT motif occurs on, the intron has significantly different RNA secondary structure. Perhaps the hairpin forming version is more ideal and is selected for somehow.
- Maybe “A” rich regions are just harder to transcribe and selected against.
While it is unclear what is driving these biases, clearly they exist and likely tell us something important about the role of STRs in transcription.
Tweet comments powered by Disqus